SEMICONSERVATIVE REPLICATION
Each original strand of DNA is used as a template for synthesis of a new strand. The new DNA according to their proposed semiconservative replication model would be a hybrid. Each new duplex would be composed of an original strand and a newly synthesized strand.
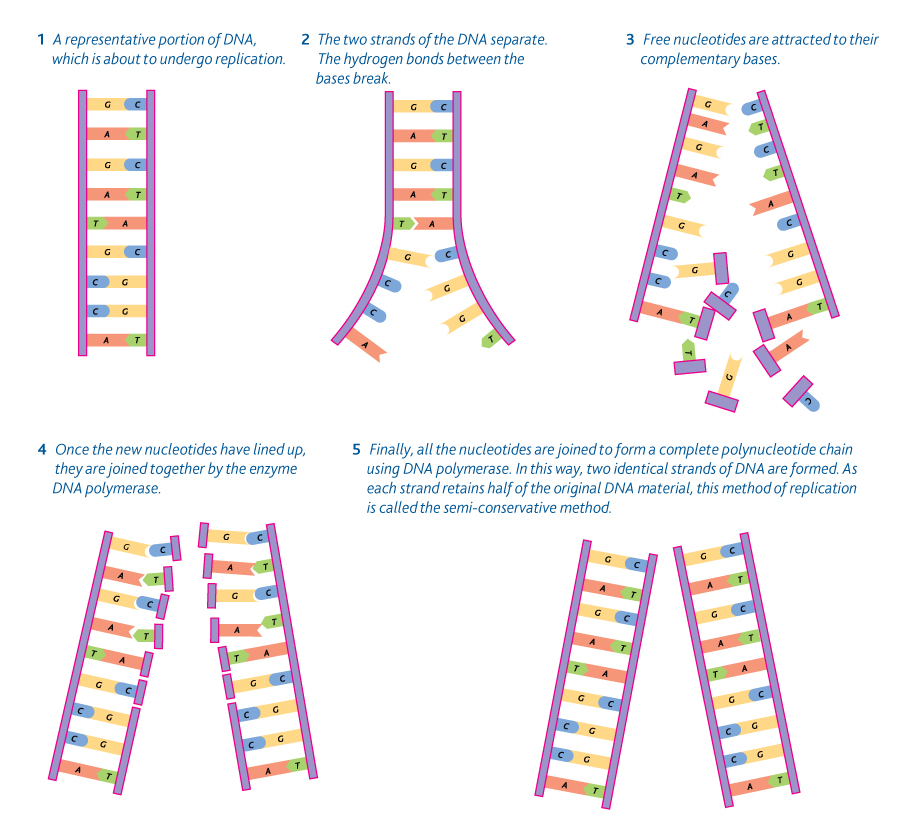 |
| Figure 1 |
Actions of DNA Polymerases
Specific requirements for the reaction process include magnesium ion for nucleotide complexation and the presence of "preformed DNA", which serves two purposes. First, the DNA acts as a template that carries the messages to be copied. The massage is read according to Watson-Crick base pairing rules . Second, the preformed DNA must have a primer segment with a free 3' -hydroxyl group. This site provides access for covalent attachment of the entering nucleotide. DNA polymerase I is unable to start a new polynucleotide chain without such a primer. In chemical terms, the reaction proceeds with nucleophilic attack by the free 3' -hydroxyl group of the primer on the entering nucleotide, which is selected and held in place by specific base pairing. Thus, elongation occurs at the 3' end with net synthesis of DNA proceeding in the 5' → 3' direction. Since the chains are antiparallel, the message in DNA template is read in the 3' → 5' direction. Energy for the reaction is supplied by release of pyrophosphate, PPi a reactive molecule that is readily hydrolyzed, with release of energy, to form two molecules of orthophosphate.
 |
| Figure 2 |
DNA Replication
Step 1:
Before DNA can be replicated, the double stranded molecule must be “unzipped” into two single strands. DNA has four bases called adenine (A), thymine (T), cytosine (C) and guanine (G) that form pairs between the two strands. Adenine only pairs with thymine and cytosine only binds with guanine. In order to unwind DNA, these interactions between base pairs must be broken. This is performed by an enzyme known as DNA helicase. DNA helicase disrupts the hydrogen bonding between base pairs to separate the strands into a Y shape known as the replication fork. This area will be the template for replication to begin.
DNA is directional in both strands, signified by a 5' and 3' end. This notation signifies which side group is attached the DNA backbone. The 5' end has a phosphate (P) group attached, while the 3' end has a hydroxyl (OH) group attached. This directionality is important for replication as it only progresses in the 5' to 3' direction. However, the replication fork is bi-directional; one strand is oriented in the 3' to 5' direction (leading strand) while the other is oriented 5' to 3' (lagging strand). The two sides are therefore replicated with two different processes to accommodate the directional difference.
Step 2:
The expose single strands of DNA must be stabilized and protected from hydrolytic cleavage of phosphodiester bonds. The single-stranded DNA binding proteins (SSB protein) perform this protective role. The now separated polynucleotide strands are used as templates for the synthesis of complementary strands.
Step 3:
The next step, initiation of DNA synthesis. bring it new twist for DNA polymerase III. The action of all DNA polymerase III, requires a primer with a free 3' -hydroxyl end. Since this is not present at the replication fork, synthesis DNA cannot begin immediately. This problem is resolved by the synthesis of a short of stretch of RNA complementary to the DNA template. RNA synthesis synthesis is catalyzed by an enzyme called primase. The action of primase is required only once for the initiation of the leading strand of DNA. On the other hand, each Okazaki fragment must be initiated by the action of primase. After the few ribonucleotides are added, DNA synthesis catalyzed by DNA polymerase III can proceed from the 3' -hydroxyl group. In later steps, the RNA primer is removed from DNA by the 5' → 3' nuclease action of DNA polymerase I.
Step 4:
Now, with the availability of the free 3' -hydroxyl group, DNA synthesis begins, catalyzed by DNA polymerases III. Both the leading strand and the lagging strand are extended in this way. The leading strand proceeds in the direction of the advancing replication fork. Synthesis of the lagging strand continues in the opposite direction until it meets the fragment previously synthesized.
Step 5:
At this time, the RNA primers are removed by the 5' → 3' nuclease action of DNA polymerase I. Small gaps remain that are filled in by the synthesizing action of DNA polymerase I
Step 6:
DNA polymerase activity can bring in all of the deoxyribonucleotide bases needed to fill in the gaps, however, the final phosphoester bond to close completely the last gap must be formed by an additional bond to close completely the last gap must be formed by an additional enzyme, DNA ligase. This enzyme catalyzes ATP-dependent phosphate ester formation between a free hydroxyl group at the 3' end of one fragment with a phosphate group 5' end of the other.
Step 4:
Now, with the availability of the free 3' -hydroxyl group, DNA synthesis begins, catalyzed by DNA polymerases III. Both the leading strand and the lagging strand are extended in this way. The leading strand proceeds in the direction of the advancing replication fork. Synthesis of the lagging strand continues in the opposite direction until it meets the fragment previously synthesized.
Step 5:
At this time, the RNA primers are removed by the 5' → 3' nuclease action of DNA polymerase I. Small gaps remain that are filled in by the synthesizing action of DNA polymerase I
Step 6:
DNA polymerase activity can bring in all of the deoxyribonucleotide bases needed to fill in the gaps, however, the final phosphoester bond to close completely the last gap must be formed by an additional bond to close completely the last gap must be formed by an additional enzyme, DNA ligase. This enzyme catalyzes ATP-dependent phosphate ester formation between a free hydroxyl group at the 3' end of one fragment with a phosphate group 5' end of the other.
No comments:
Post a Comment